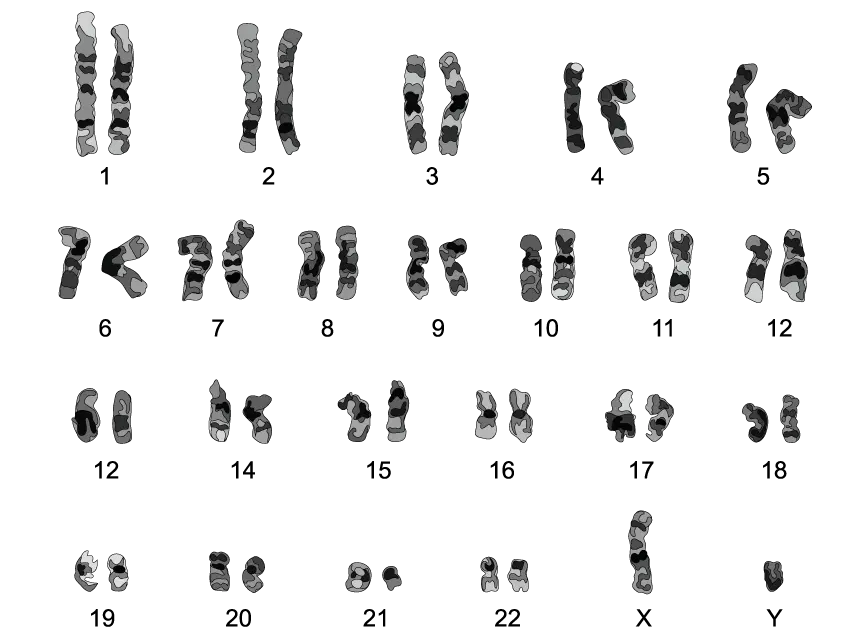
The following diagram shows the karyogram of an individual.

(i) Identify the sex of this individual.
[1]
(ii) State a reason for your answer in part i).
[1]
Cells in metaphase of mitosis were used to construct the karyogram from part (a).
Explain the reason for this.
List two characteristics of the chromosomes that are used to arrange them in a karyogram.
Apart from sex determination, state one other use of studying the karyotype of an individual.
Was this exam question helpful?